Saving and loading geometries
In many cases it is beneficial to save geometries into a file for further use. For example the generation of 3D geometry data from STL-files consumes a lot of CUP time. For a series of simulation it is useful to generate the geometry data only once. Similarly if one calculates large simulations on a cluster computer, it may be useful to still do plots on a local desktop. Even in the case FuncSolids are used, the geometry data output from IBSimu simulations can contain all information necessary for making plots and/or analysis.
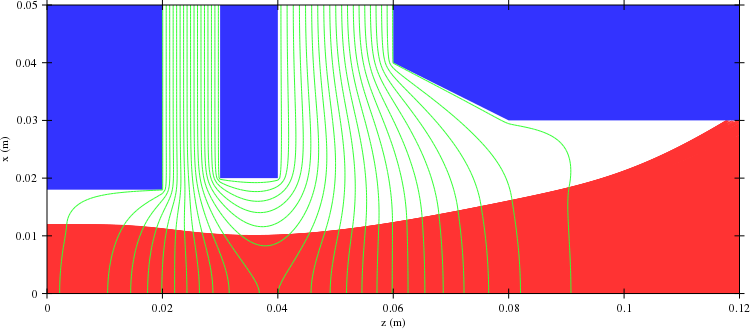
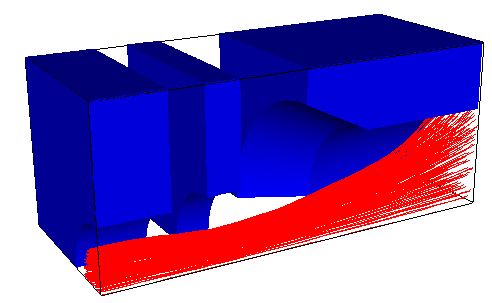
Here is a simple example of a two-part 3D simulation. The first program builds a geometry using FuncSolids with build_mesh() and saves it to geom.dat. This data includes the following information:
- Dimensions of the mesh
- Boundary conditions
- Inclusion of each mesh node. Information if a node is vacuum or inside a solid.
- Distance information for nodes near solids. If a node is within one mesh unit of a solid the distance to the solid surface is saved.
The second program loads the geometry from the file and calls geom.build_surface(). This creates a surface triangulation representation of the original solid surfaces using so-called marching cubes algorithm. The triangulation is a fast operation, which is why the triangle information is not saved into the geometry data file. The particle iteration uses solid data by default for collision detection. In cases like this, where the original solid data is not available one can use the surface triangulation data for collision detection. This is enabled with set_surface_collision( true ). The surface triangulation data is also used for 3D rendered geometry views. The 2D geometry plots are made using only inclusion and near solid data (3 and 4 in the list above).
Please note that at the time of writing no surface collision algorithm has been made for 2D simulations. Therefore the above only applies to 3D simulations.
Saving program (save.cpp):
#include "epot_bicgstabsolver.hpp"
#include "geometry.hpp"
#include "func_solid.hpp"
#include "epot_efield.hpp"
#include "meshvectorfield.hpp"
#include "particledatabase.hpp"
#include "geomplotter.hpp"
#include "ibsimu.hpp"
#include "error.hpp"
bool solid1( double x, double y, double z )
{
double r = sqrt(x*x+y*y);
return( z <= 0.02 && r >= 0.018 );
}
bool solid2( double x, double y, double z )
{
double r = sqrt(x*x+y*y);
return( z >= 0.03 && z <= 0.04 && r >= 0.02 );
}
bool solid3( double x, double y, double z )
{
double r = sqrt(x*x+y*y);
return( z >= 0.06 && r >= 0.03 && r >= 0.07 - 0.5*z );
}
void simu( void )
{
Geometry geom( MODE_3D, Int3D(101,101,241), Vec3D(0,0,0), 5e-4 );
Solid *s1 = new FuncSolid( solid1 );
geom.set_solid( 7, s1 );
Solid *s2 = new FuncSolid( solid2 );
geom.set_solid( 8, s2 );
Solid *s3 = new FuncSolid( solid3 );
geom.set_solid( 9, s3 );
geom.set_boundary( 1, Bound(BOUND_NEUMANN, 0.0 ) );
geom.set_boundary( 2, Bound(BOUND_NEUMANN, 0.0 ) );
geom.set_boundary( 3, Bound(BOUND_NEUMANN, 0.0 ) );
geom.set_boundary( 4, Bound(BOUND_NEUMANN, 0.0 ) );
geom.set_boundary( 5, Bound(BOUND_DIRICHLET, -3.0e3) );
geom.set_boundary( 6, Bound(BOUND_DIRICHLET, -1.0e3) );
geom.set_boundary( 7, Bound(BOUND_DIRICHLET, -3.0e3) );
geom.set_boundary( 8, Bound(BOUND_DIRICHLET, -14.0e3) );
geom.set_boundary( 9, Bound(BOUND_DIRICHLET, -1.0e3) );
geom.build_mesh();
geom.save( "geom.dat" );
}
int main( int argc, char **argv )
{
try {
ibsimu.set_message_threshold( MSG_VERBOSE, 1 );
ibsimu.set_thread_count( 4 );
simu();
} catch ( Error e ) {
e.print_error_message( ibsimu.message( 0 ) );
exit( 1 );
}
return( 0 );
}
Loading program (load.cpp):
#include "epot_bicgstabsolver.hpp"
#include "geometry.hpp"
#include "func_solid.hpp"
#include "epot_efield.hpp"
#include "meshvectorfield.hpp"
#include "particledatabase.hpp"
#include "geomplotter.hpp"
#include "gtkplotter.hpp"
#include "ibsimu.hpp"
#include "error.hpp"
void simu( int argc, char **argv )
{
std::string geom_fn = "geom.dat";
std::ifstream is_geom( geom_fn.c_str() );
if( !is_geom.good() )
throw( Error( ERROR_LOCATION, (std::string)"couldn\'t open file \'" + geom_fn + "\'" ) );
Geometry geom( is_geom );
is_geom.close();
geom.build_surface();
EpotField epot( geom );
MeshScalarField scharge( geom );
MeshVectorField bfield;
EpotEfield efield( epot );
field_extrpl_e efldextrpl[6] = { FIELD_EXTRAPOLATE, FIELD_EXTRAPOLATE,
FIELD_SYMMETRIC_POTENTIAL, FIELD_EXTRAPOLATE,
FIELD_EXTRAPOLATE, FIELD_EXTRAPOLATE };
efield.set_extrapolation( efldextrpl );
EpotBiCGSTABSolver solver( geom );
ParticleDataBase3D pdb( geom );
bool pmirror[6] = { false, false, true, false, false, false };
pdb.set_mirror( pmirror );
pdb.set_surface_collision( true );
for( size_t i = 0; i < 5; i++ ) {
solver.solve( epot, scharge );
efield.recalculate();
pdb.clear();
pdb.add_cylindrical_beam_with_energy( 100000, 50.0, 1.0, 1.0,
3.0e3, 0.0, 0.0,
Vec3D(0,0,0),
Vec3D(1,0,0),
Vec3D(0,1,0),
0.012 );
pdb.iterate_trajectories( scharge, efield, bfield );
}
GeomPlotter geomplotter( geom );
geomplotter.set_size( 750, 750 );
geomplotter.set_epot( &epot );
geomplotter.set_particle_database( &pdb );
geomplotter.set_view( VIEW_ZX, 0 );
geomplotter.plot_png( "plot_zx.png" );
if( true ) {
MeshScalarField tdens( geom );
pdb.build_trajectory_density_field( tdens );
GTKPlotter plotter( &argc, &argv );
plotter.set_geometry( &geom );
plotter.set_epot( &epot );
plotter.set_bfield( &bfield );
plotter.set_efield( &efield );
plotter.set_scharge( &scharge );
plotter.set_trajdens( &tdens );
plotter.set_particledatabase( &pdb );
plotter.new_geometry_plot_window();
plotter.run();
}
}
int main( int argc, char **argv )
{
try {
ibsimu.set_message_threshold( MSG_VERBOSE, 1 );
ibsimu.set_thread_count( 4 );
simu( argc, argv );
} catch ( Error e ) {
e.print_error_message( ibsimu.message( 0 ) );
exit( 1 );
}
return( 0 );
}
The files:
Plots from load.cpp: